
- Cluego cytoscape tutorial update#
- Cluego cytoscape tutorial driver#
- Cluego cytoscape tutorial code#
- Cluego cytoscape tutorial download#
hierarchical, select the corresponding option from the Cytoscape Visualization menu. For more significant p-values, the node color gets increasingly more orange (see also the Color Legend panel). Yellow nodes represent GO categories that are overrepresented at the significance level. Uncolored nodes are not overrepresented, but they are the parents of overrepresented categories further down. Finally, a visualization of the overrepresented GO categories is created in Cytoscape. The program will inform you of its progress while parsing the annotations and calculating the tests, corrections and layout. Finally, select a directory to save the output file in (the file will be named test.bgo if you filled in test as a cluster name), and press Start BiNGO.
Cluego cytoscape tutorial code#
We want to consider all evidence codes, so don't fill in anything in the evidence code box. Select GO_Biological_Process from the ontology list, and Saccharomyces cerevisiae from the organism list. We're interested in assessing the overrepresentation of functional categories in our cluster with respect to the whole yeast genome, which is why we choose the Complete Annotation as the reference set. Since we only want to visualize those GO categories that are overrepresented after multiple testing correction, and their parents in the GO hierarchy, select the corresponding visualization option. Then select a statistical test (the Hypergeometric Test is exact and equivalent to an exact Fisher test, the Binomial Test is less accurate but quicker) and a multiple testing correction (we recommend Benjamini & Hochberg's FDR correction, the Bonferroni correction will be too conservative in most cases), and choose a significance level, e.g. The corresponding boxes are checked accordingly by default. We want to assess overrepresentation of GO categories, and we want to visualize the results in Cytoscape. Check the box Get Cluster from Network (see below for an example with text input). This name will be used for creating the output file and the visualization of the results in Cytoscape. Start by filling in a name for your cluster. Chromosomal locations can be automatically updated (NCBI).The BiNGO Settings panel pops up. Mapping on chromosomal locations is enabled. Reactome data split into Reaction and Pathways. Multiple ontologies with separate OBO files. Organism creation date included for new organisms New organisms included upon user request
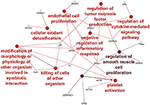
New analysis suggestions and explanations as help button included
Cluego cytoscape tutorial update#
Improved network update behavior and speed Additional iterative merging of functional groups for large networks
Cluego cytoscape tutorial download#
InterPro download was not working due to a change of the source data link
Cluego cytoscape tutorial driver#
In certain graphic card driver versions a stricter usage of data types is needed Overlapping terms were included more than once in a group Percentage of not found genes are now reported to unique genes, not to the initial uploaded ids Correction of the stats displayed in the ClueGO log. ClueGO functions are now REST enabled, and allow the integration of ClueGO in analytical pipelines. To extend it's functionality get as well CluePedia.ĬlueGO visualizes also terms and pathways resulted from other enrichment analyses as functionally grouped networks.Ĭytoscape Automation enables scientific workflows written in many languages through the CyREST. ClueGO is easy updatable with the newest files from Gene Ontology, KEGG, WikiPathways and Reactome. The significance of the terms and groups is automatically calculated. ClueGO charts are underlying the specificity and the common aspects of the biological role. On the network, the node colour can be switched between functional groups and clusters distribution. The ClueGO network is created with kappa statistics and reflects the relationships between the terms based on the similarity of their associated genes. The related terms which share similar associated genes can be fused to reduce redundancy. From the ontology sources used, the terms are selected by different filter criteria. ClueGO performs single cluster analysis and comparison of several clusters (lists of genes). The type of identifiers supported can be easily extended by the user. The identifiers can be uploaded from a text file or interactively from a network of Cytoscape. ClueGO is a Cytoscape plug-in that visualizes the non-redundant biological terms for large clusters of genes in a functionally grouped network.


 0 kommentar(er)
0 kommentar(er)
